INF| Schütte/ Ludwig
Information Management and Information Infrastructure in the Collaborative Research Center
The aim of project INF is to provide a central hub that supports all researchers of the CRC 1449 in the organization of collaboration (e.g. exchange of sample protocols or experiment design) as well as in the exchange, processing, analysis, and archiving of scientific data. This project is supported by and in collaboration with the Zuse Institute Berlin (ZIB) that provides part of the critical infrastructure. In addition to providing the necessary technical basis for this, project INF also creates an awareness of the importance and the requirements for the research data management. As part of NFDI4Chem, we are actively working on the implementation of tools and standards that support FAIR data handling in chemical research.
Infrastructure provided by INF:
- Document Exchange: To share documents and smaller data sets an internal FU-Box and Nextcloud are provided to allow for seamless collaboration and mobile working.
- Booking Plattform: Establishment of OpenIRIS as a uniform resource and booking platform shared at the different locations to give all researchers within the CRC a better overview of the available measurements and analysis resources and to simplify direct booking of instruments.
- Data Storage: Provision of ~1 PB Storage system with long-term archiving of the data for at least 10 years to follow best scientific practices.
- Data Enrichment: We are working on a support infrastructure for the enrichment of the primary data with metadata to improve findability and reusability.
- Data Sharing: We are currently working on an extension of the data storage with a webbased share point for the exchange of scientific research data within the CRC. A documentation of the preliminary version can be found here.
- Electronic Lab Notebook: All researchers of the CRC are offered a license for the ELN Labfolder to document and organize their research projects. We are currently testing the open-source ELNs eLabFTW and Chemotion in cooperation with the Berlin University Alliance (BUA, CARDS TP4). If you are interested in an account, reach out to Dr. Yannic Kerkhoff (kerkhoff@zib.de).
Image and data analysis tools developed by INF:
A collection of data analysis tools developed by INF can be found at Zenodo. Developed and established tools are offered as analysis service by Project Z01 Bioanalytical Core Unit. All participants of this CRC are also invited to upload their software and data sets in the dedicated CRC1449 Zenodo Community.
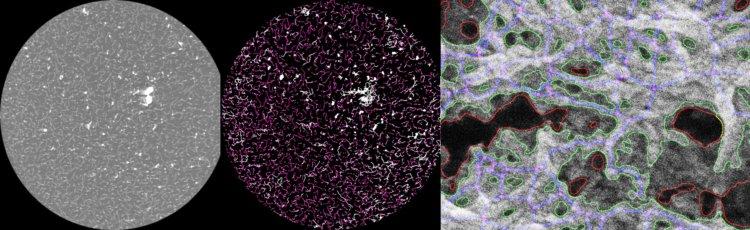
Representation of cryo-EM mucus-inspired polymer fiber analysis (left) and AI-based SEM-imaged human mucus network analysis (right) developed by INF.
Scientific chatbot hosted at ZIB:
Together with the ZIB we set up a Scientific Chatbot that is based on Open WebUI and hosted at ZIB. It allows for support in e.g. literature research, scientific writing, and coding while keeping all your research data safe at our own servers. We are currently developing a portfolio of specialized AI Advisors for scalable support and consulting in areas such as Bioimage Analysis, Biostatistiscs, FAIR Data Handling, Manuscript Reviewing and more. A guide for the usage of the chatbot can be found here. An early version can be accessed in the FUB net or via VPN with a zedat account here: ollama-bcp-zib.de

Use case example of the Scientific Chatbot: CRC members can have their manuscripts reviewed by an AI system to receive feedback on strengths, weaknesses, improvements, and suitable follow-up experiments. Their unpublished data remains safe on the servers of the ZIB.
- To access the chatbot your zedat account must be activated in the MI-Portal
For external users: People outside of FUB that want to use our infrastructure (e.g., share point, ELNs, chatbot, etc.) need a FU project account, that can be requested from Dr. Wiebke Fischer.
Software and data training provided by INF:

CRC-members from TU and Charité learn how to train and apply machine-learning tools to analyze human mucus samples. F.l.t.r: Merten Strehlau (TUB, A01), Christine Wong (Charité, A01), Aditi Löwe (Charité, C04), Pelin Sungur (Charité, B01), Ketaki Sandu (Charité, B04), and Yannic Kerkhoff (FUB/ZIB, INF&Z01).
- A collection of training videos for image analysis with Fiji can be found here.
- The documentation including tutorial videos for the AI-supported quantification of SEM-imaged network structures can be found here.
Selected CRC publications with essential contributions from INF:
Efficient AI-supported quantification of network structures in SEM imaged human mucus. Y. Kerkhoff, H. Rulff, P. W. Mönch, C. Gutwein, A. Loewe, S. Timm, R. M. Urbantat, K. Ludwig, R. Haag, M. Gradzielski, M. A. Mall, B. Siegmund, M. Ochs. chemRxiv. 2025, 10.26434/chemrxiv-2025-qz5bj:
INF developed an AI-based method for the fast and reliable quantification of SEM-imaged mucus network structures.
Polysialosides Outperform Sulfated Analogs for Binding with SARS-CoV-2. V. Khatri, N. Boback, H. Abdelwahab, D. Niemeyer, T. M. Palmer, A. K. Sahoo, Y. Kerkhoff, K. Ludwig, J. Heinze, D. Balci, J. Trimpert, R. Haag, T. L. Povolotsky, R. R. Netz, C. Drosten, D. C. Lauster, S. Bhatia. Small. 2025, 2500719:
INF trained a small neural network to detect polymer-based nanoparticles and viruses on cryo-ET images to visualize binding events.
Supramolecular Architectures of Dendritic Polymers Provide Irreversible Inhibitor to Block Viral Infection. E. Mohammadifar, M. Gasbarri, M. Dimde, C. Nie, H. Wang, T. L. Povolotsky, Y. Kerkhoff, D. Desmecht, S. Prevost, T. Zemb, K. Ludwig, F. Stellacci, R. Haag. Adv. Mater. 2024, 2408294:
INF trained a small neural network to detect different nanoparticles on cryo-TEM images to segment polymeric nanoparticles and viruses to elucidate 3D binding conformations.
Mucus-Inspired Self-Healing Hydrogels: A Protective Barrier for Cells against Viral Infection. R. Bej, C. Alfred Stevens, C. Nie, K. Ludwig, G. Degen, Y. Kerkhoff, M. Pigaleva, J. M. Adler, N. A. Bustos, T. M. Page, J. Trimpert, S. Block, B. B. Kaufer, K. Ribbeck, R. Haag. Adv. Mater., 2024, 36(32):e2401745:
INF developed a machine-learning assisted ImageJ macro that allows for the 3D reconstruction and analysis of polymers fibers from cryo-EM images to validate rationally designed inhibitor properties.
Mucin-Inspired Polymeric Fibers for Herpes Simplex Virus Type 1 Inhibition. J. Arenhoevel, A.-C. Schmitt, Y. Kerkhoff, V. Ahmadi, E. Quaas, K. Ludwig, K. Achazi, C. Nie, R. Bej, R. Haag. Macromol. Biosci. 2024, 24, 2400120:
INF developed a machine-learning based ImageJ plugin for the automatic and reproducible analysis of plaque (reduction) assays from well plate images.
Comprehensive Characterization of the Viscoelastic Properties of Bovine Submaxillary Mucin (BSM) Hydrogels and the Effect of Additives. H. Rulff, R. F. Schmidt, L.-F. Wei, K. Fentker, Y. Kerkhoff, P. Mertins, M. A. Mall, D. Lauster, M. Gradzielski. Biomacromolecules, 2024, 25, 7, 4014–4029:
INF developed an AI-assisted analysis scheme for the automatic analysis of porous structures on Scanning Electrone Microscopy (SEM) images for the characterization of mucus properties.
Polysulfates Block SARS-CoV-2 Uptake through Electrostatic Interactions. C. Nie, P. Pouyan, D. Lauster, J. Trimpert, Y. Kerkhoff, G. P. Szekeres, M. Wallert, S. Block, A. K. Sahoo, J. Dernedde, K. Pagel, B. B. Kaufer, R. R. Netz, M. Ballauff, R. Haag, Angew. Chem. Int. Ed. 2021, 60, 15870:
INF developed a script for the high-throughput detection of virus-particles on cells to quantify the influence of sulfated polymer inhibitors on the progression of viral infections.
Contact:
If you have any questions please reach out to Dr. Yannic Kerkhoff.